
NSF ABI Innovation: A New Automated Data Integration, Annotations, and Interaction
Network Inference System for Analyzing Drosophila Gene Expression
National Science Foundation Award Number: 1356628
Contact Information
Heng Huang, PI
Chris Ding, Co-PI
Computer Science and Engineering Department
University of Texas at Arlington
500 UTA Blvd.
Arlington, TX 76019 U.S.A.
Project Award Information
- Click here to see the Award information on NSF website.
Project Summary
Large-scale in situ hybridization (ISH) screens are providing an abundance of spatio-temporal patterns of gene expression data that is valuable for understanding the mechanisms of gene regulation. Drosophila gene expression pattern images enable the integration of spatial expression patterns with other genomic datasets that link regulator with their downstream targets. This project addresses the computational challenges in analyzing Drosophila gene expression patterns by leveraging a new bioinformatics software system, and focuses on designing principled bioinformatics and computational biology algorithms and tools that will integrate multi-modal spatial patterns of gene expression for Drosophila embryos developmental stage recognition and anatomical ontology term annotation, and infer the gene interaction network to generate a more comprehensive picture of genome function and interaction. The bioinformatics methods resulted from the project activities are directly transformable to a variety of fields such as biomedical science and engineering, system biology, clinical pathology, oncology, and pharmaceutics.
This project investigates three challenging problems for studying the Drosophila Embryo ISH Images via innovative bioinformatics algorithms: 1) the sparse multi-dimensional feature learning method to integrate the multimodal spatial gene expression patterns for annotating Drosophila ISH images, 2) the heterogeneous multi-task learning models using high-order relational graph to jointly recognize the developmental stages and annotate anatomical ontology terms, 3) the embedded sparse representation algorithm to infer the gene interaction network. It is innovative to apply structured sparse learning, multi-task learning, and high-order relational graph models to Drosophila gene expression patterns analysis that holds great promise for scientific investigations into the fundamental principles of animal development. The developed algorithms and tools are expected to help knowledge discovery for applications in broader scientific and biological domains with massive high-dimensional and heterogonous data sets. This project facilitates the development of novel educational tools to enhance several current courses at University of Texas at Arlington. The PIs are engaging the minority students and under-served populations in research activities to give them a better exposure to cutting-edge science research.
Publications and Products
Nugget 1. Multi-Dimensional Visual Descriptors Integration Algorithm for Drosophila Gene Expression Patterns Anatomical Annotations:
To facilitate the search and comparison of Drosophila gene expression patterns during Drosophila embryogenesis, it is highly desirable to annotate the tissue-level anatomical ontology terms for ISH images. In ISH image annotations, the image content representation is crucial to achieve satisfactory results. However, existing methods mainly focus on improving the classification algorithms and only using simple visual descriptor. We propose a novel structured sparsity-inducing norms based feature learning model to integrate the multi-dimensional visual descriptors for Drosophila gene expression patterns annotations. The new mixed norms are designed to learn the importance of different features from both local and global point of views. We successfully integrate six widely used visual descriptors to annotate the Drosophila gene expression patterns from the lateral, dorsal, and ventral views. The empirical results show that the proposed new method can effectively integrate different visual descriptors, and consistently outperforms related methods using the concatenated visual descriptors.
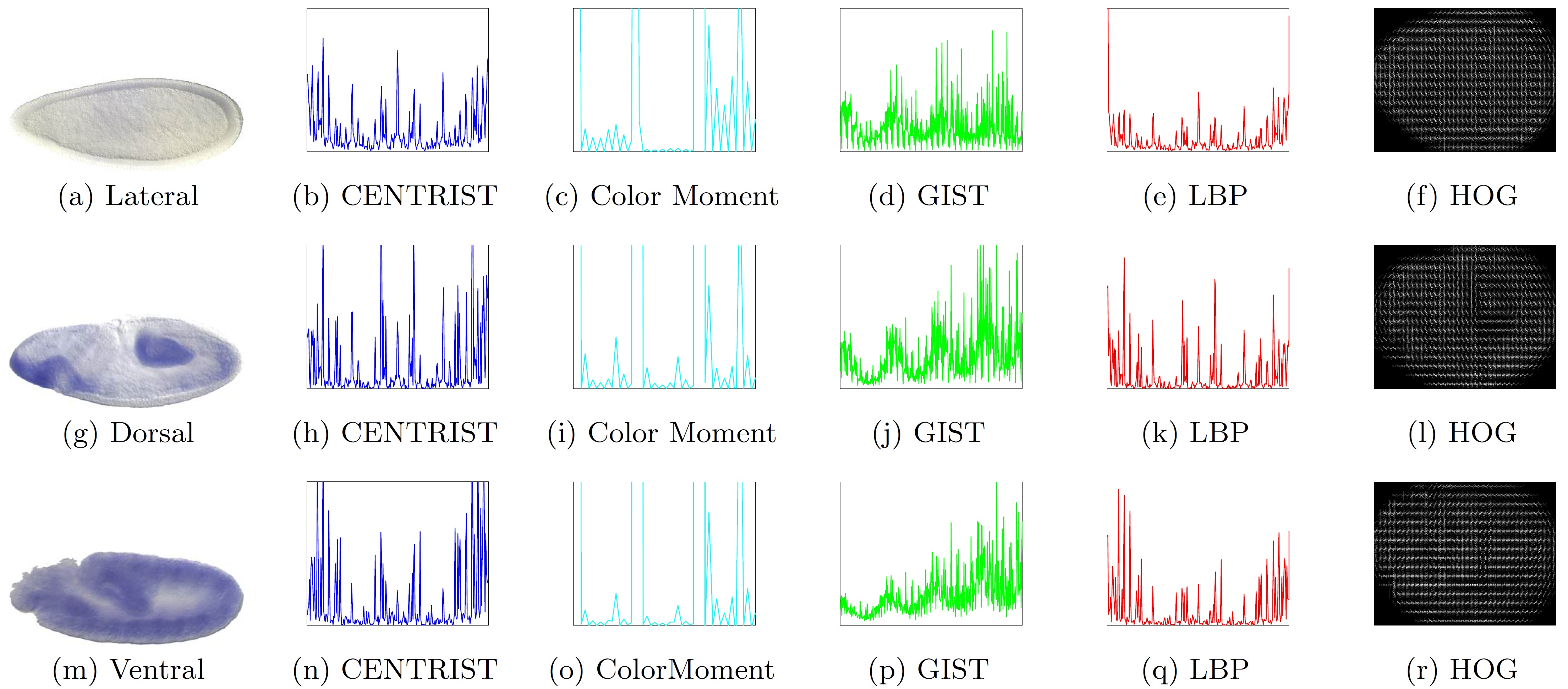
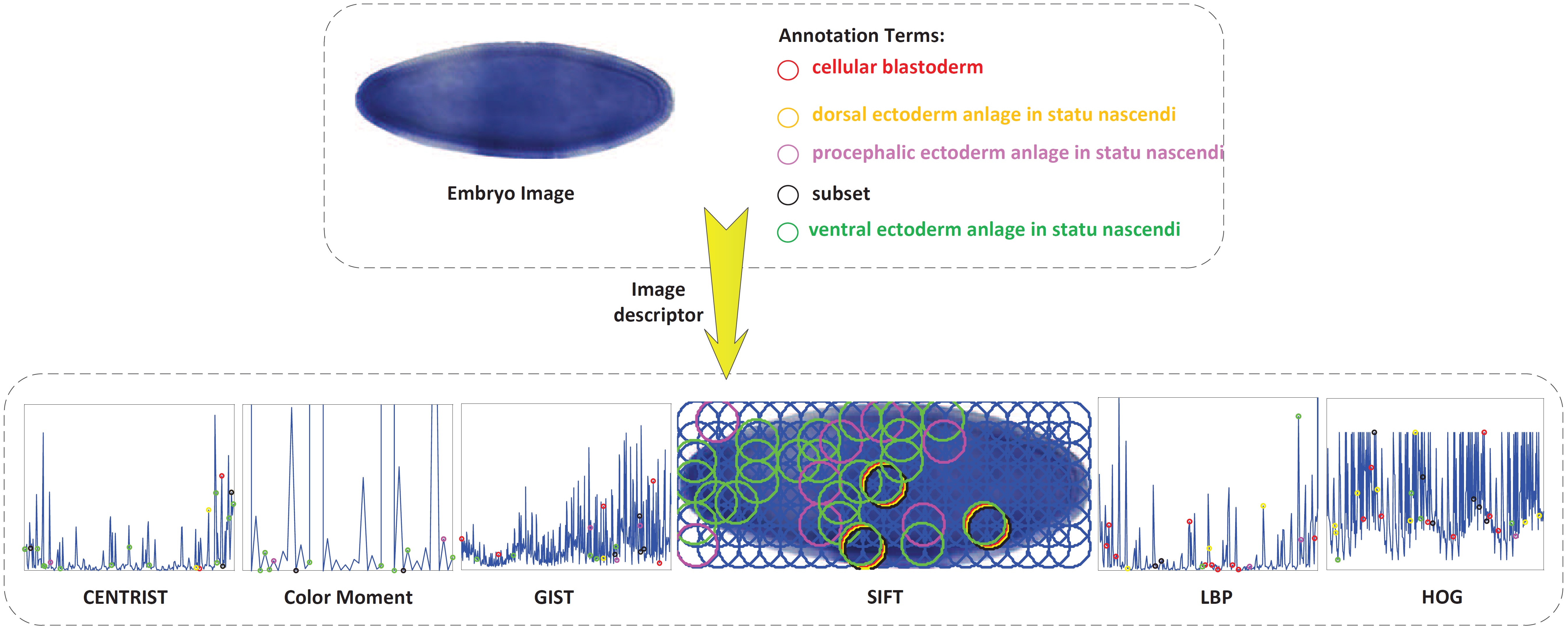
This work was published in KDD 2015:
Hongchang Gao, Lin Yan, Weidong Cai, Heng Huang. Anatomical Annotations for Drosophila Gene Expression Patterns via Multi-Dimensional Visual Descriptors Integration. 21th ACM SIGKDD Conference on Knowledge Discovery and Data Mining Conference (KDD 2015), pp. 339-348.
Nugget 2. Drosophila Gene Expression Pattern Annotations via Multi-Instance Biological Relevance Learning:
Recent developments in biology have produced a large number of gene expression patterns, many of which have been annotated textually with anatomical and developmental terms. These terms spatially correspond to local regions of the images, which are attached collectively to groups of images. Because one does not know which term is assigned to which region of which image in the group, the developmental stage classification and anatomical term annotation turn out to be a multi-instance learning (MIL) problem, which considers input as bags of instances and labels are assigned to the bags. Most existing MIL methods routinely use the Bag-to-Bag (B2B) distances, which, however, are often computationally expensive and may not truly reflect the similarities between the anatomical and developmental terms. We approach the MIL problem from a new perspective using the Class-to-Bag (C2B) distances, which directly assesses the relations between annotation terms and image panels. Taking into account the two challenging properties of multi-instance gene expression data, high heterogeneity and weak label association, we computes the C2B distance by introducing class specific distance metrics and locally adaptive significance coefficients. We apply our new approach to automatic gene expression pattern classification and annotation on the Drosophila melanogaster species. Our experiments demonstrated the effectiveness of the new multi-instance learning method.
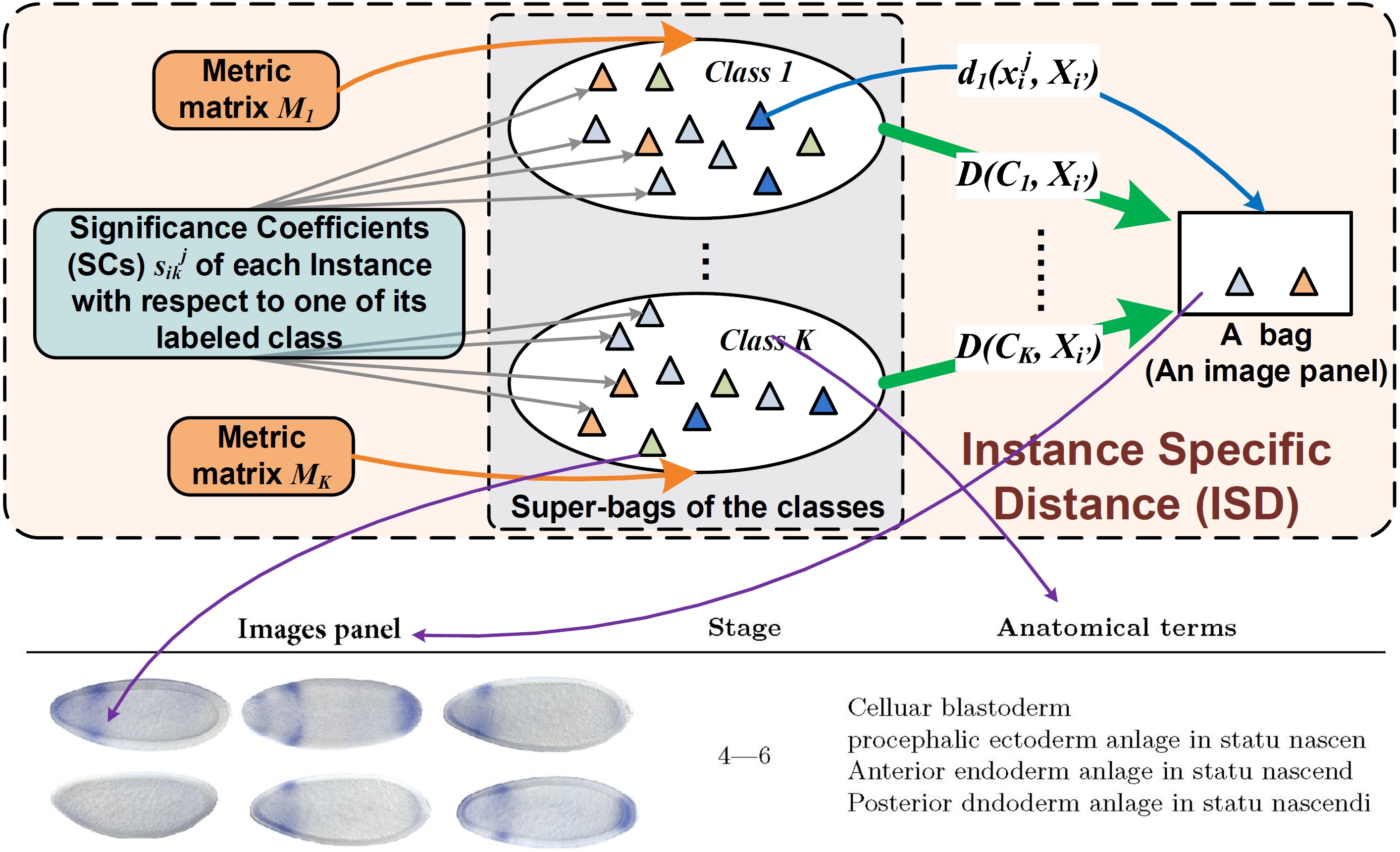
This work was published in AAAI 2016:
Hua Wang, Cheng Deng, Hao Zhang, Xinbo Gao, Heng Huang. Learning Biological Relevance of Drosophila Embryos for Drosophila Gene Expression Pattern Annotations. Thirtieth AAAI Conference on Artificial Intelligence (AAAI 2016) , pp. 1324-1330.
Some Related Publications:
Lei Du, Heng Huang, Jingwen Yan, Sungeun Kim, Shannon L. Risacher, Mark Inlow, Jason H. Moore, Andrew J. Saykin, Li Shen, ADNI. Structured Sparse Canonical Correlation Analysis for Brain Imaging Genetics: An Improved GraphNet Method. Bioinformatics, 32(10):1544-51.De Wang, Feiping Nie, Heng Huang. Global Redundancy Minimization for Feature Ranking. IEEE Transactions on Knowledge and Data Engineering (TKDE), 27(10), pp. 2743-2755, 2015.
Xiaoqian Wang, Feiping Nie, Heng Huang. Structured Doubly Stochastic Matrix for Graph Based Clustering. 22nd ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD 2016), accepted to appear.
Zhouyuan Huo, Feiping Nie, Heng Huang. Robust and Effective Metric Learning Using Capped Trace Norm. 22nd ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD 2016), accepted to appear.
Wenhao Jiang, Hongchang Gao, Fu-Lai Korris Chung, Heng Huang. L2,1-Norm Stacked Robust Autoencoders for Domain Adaptation. Thirtieth AAAI Conference on Artificial Intelligence (AAAI 2016) , pp. 1723-1729.
Hua Wang, Cheng Deng, Hao Zhang, Xinbo Gao, Heng Huang. Learning Biological Relevance of Drosophila Embryos for Drosophila Gene Expression Pattern Annotations. Thirtieth AAAI Conference on Artificial Intelligence (AAAI 2016) , pp. 1324-1330.
Hongchang Gao, Feiping Nie, Heng Huang. Multi-View Subspace Clustering. International Conference on Computer Vision (ICCV 2015), pp. 4238-4246.
Hongchang Gao, Chengtao Cai, Jingwen Yan, Lin Yan, Joaquin Goni Cortes, Yang Wang, Feiping Nie, John West, Andrew Saykin, Li Shen, Heng Huang. Identifying Connectome Module Patterns via New Balanced Multi-Graph Normalized Cut. 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2015), pp. 169-176.
Yang Song, Weidong Cai, Heng Huang, Yun Zhou, Yue Wang, David Dagan Feng. Locality constrained Subcluster Representation Ensemble for Lung Image Classification. Medical Image Analysis (MIA), 22(1), pp. 102-113, 2015.
Hua Wang, Feiping Nie, Heng Huang. Large-Scale Cross-Language Web Page Classification via Dual Knowledge Transfer Using Fast Nonnegative Matrix Tri-Factorization. ACM Transactions on Knowledge Discovery from Data (TKDD), 10(1), pp. 1:1-1:29, 2015.
Yang Song, Weidong Cai, Heng Huang, Yun Zhou, David Dagan Feng, Yue Wang, Michael J. Fulham, Mei Chen. Large Margin Local Estimate with Applications to Medical Image Classification. IEEE Transactions on Medical Imaging (TMI), 34(6), pp. 1362-1377, 2015.
Hua Wang, Heng Huang, Chris Ding. Correlated Protein Function Prediction via Maximization of Data Knowledge Consistency. Journal of Computational Biology (JCB), 22(6), pp. 546-562, 2015.
Hongchang Gao, Lin Yan, Weidong Cai, Heng Huang. Anatomical Annotations for Drosophila Gene Expression Patterns via Multi-Dimensional Visual Descriptors Integration. 21th ACM SIGKDD Conference on Knowledge Discovery and Data Mining Conference (KDD 2015), pp. 339-348.
Xiaoqian Wang, Yun Liu, Feiping Nie, Heng Huang. Discriminative Unsupervised Dimensionality Reduction. Twenty-Fourth International Joint Conferences on Artificial Intelligence (IJCAI 2015), pp. 3925-3931.
Wenhao Jiang, Feiping Nie, Heng Huang. Robust Dictionary Learning with Capped L1 Norm. Twenty-Fourth International Joint Conferences on Artificial Intelligence (IJCAI 2015), pp. 3590-3596.
Yang Song, Weidong Cai, Qing Li, Dagan Feng, Heng Huang. Fusing Subcategory Probabilities for Texture Classification. Twenty-Eighth IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2015), pp. 4409-4417.
Shuai Zheng, Xiao Cai, Chris Ding, Feiping Nie, Heng Huang. A Closed Form Solution to Multiview Low-rank Regression. Twenty-Ninth AAAI Conference on Artificial Intelligence (AAAI 2015), pp. 1973-1979.
List of any software being distributed by project
Software 1: Multi-Dimensional Visual Descriptors Integration Algorithm for Drosophila Gene Expression Patterns Anatomical Annotations:
Click to Download
Software 2: Drosophila Gene Expression Pattern Annotations via Multi-Instance Biological Relevance Learning:
Click to Download
Software 3: Robust and Effective Metric Learning Using Capped Trace Norm:
Click to Download
Software 4: Global Redundancy Minimization for Feature Ranking:
Click to Download
List of any datasets being distributed from project
All data used in this project were downloaded from Berkeley Drosophila Genome Project (BDGP). Please download the data from the original database at: http://fruitfly.org/index.html
Other Related Links