
NSF-III: Medium: Collaborative Research: Robust Large-Scale Electronic Medical Record Data Mining Framework to
Conduct Risk Stratification for Personalized Intervention
National Science Foundation Award Number: 1302675
Contact Information
Heng Huang, PI
Computer Science and Engineering Department
University of Texas at Arlington
500 UTA Blvd.
Arlington, TX 76019 U.S.A.
Song Zhang, PI
Clinical Sciences Department
University of Texas Southwestern Medical Center at Dallas
Jing Cao, PI
Statistics Department
Southern Methodist University
Project Award Information
- Click here to see the Award information on NSF website.
Project Summary
The increasingly large amounts of Electronic Medical Record (EMR) data provide us unprecedented opportunities for EMR data mining to enhance health care experiences for personalized intervention, improve different diseases risk stratifications, and facilitate understanding about disease and appropriate treatment. To solve the key and challenging problems in mining such large-scale heterogeneous EMRs, we develop a novel robust machine learning framework targeting to explore the following research tasks. First, we develop new computational tools to automate the EMRs processing, including missing values imputation by a new robust rank-k matrix completion method and unstructured free-text EMRs annotations via multi-label multi-instance learning model. Second, we investigate the new sparse multi-view learning model to integrate heterogeneous EMRs for predicting the readmission risk of Heart Failure (HF) patients and providing support to personalized intervention. Third, to identify the longitudinal patterns, we design novel high-order multi-task feature learning and classification methods. Fourth, we build the nonparametric Bayesian model for predicting the event time outcomes of the HF patients readmission. The developed sparse multi-view feature learning and robust multi-task longitudinal pattern finding frameworks enable new computational applications in a large number of research areas.
The proposed research work is innovative and crucial not only to addressing emerging EMRs applications, but also to facilitating machine learning and data mining techniques. We make the developed computational methods and tools online, available to the public. These methods and tools are expected to impact other EMR and public health research and enable investigators working on EMRs to effectively test risk prediction hypothesis. The developed algorithms and tools are expected to help knowledge extraction for applications in broader scientific and biomedical domains with massive high-dimensional and heterogonous data sets. This project will facilitate the development of novel educational tools to enhance several current courses at UT Arlington, UTSW, and SMU. The PIs are engaging the minority students and under-served populations in research activities to give them a better exposure to cutting-edge science research.
Publications and Products
Nugget 1. Missing data imputation method for longitudinal data completion:
In the longitudinal electronic medical records, because many entries are measured and changed every 24 hours, e.g. cholesterol measures, blood pressure, it is necessary to impute the missing data using the time-series data. We developed a novel spatially-temporally consistent tensor completion method to utilize both spatial and temporal information of the longitudinal data. We introduce a new smoothness regularization to utilize the content continuity within data. Besides using the new smoothness regularization to keep the temporal consistency, we also minimize the trace norm of each individual time stage data to make use of the spatial correlations among data elements.

This work was published in AAAI 2014.
Nugget 2. Natural language processing based document abstractive summarization framework:
In electronic medical record data, there are unstructured data, such as physician notes, and it is challenging to extract useful information from such unstructured data. While much work has been done in the area of extractive summarization, there has been limited study in abstractive summarization as this is much harder to achieve. We propose a new weakly supervised abstractive summarization framework using pattern based approaches. Our system first generates meaningful patterns from sentences. Then, in order to precisely cluster patterns, we propose a novel semi-supervised pattern learning algorithm that leverages a hand-crafted list of topic-relevant keywords, which are the only weakly supervised information used by our framework to generate aspect-oriented summarization. After that, our system generates new patterns by fusing existing patterns and selecting top ranked new patterns via the recurrent neural network language model. Finally, we introduce a new pattern based surface realization algorithm to generate abstractive summaries. Our new abstractive summarization technique can help users to extract useful knowledge from the unstructured electronic medical record data, such as physician notes. The summarizations can also be converted as word vectors for risk prediction.
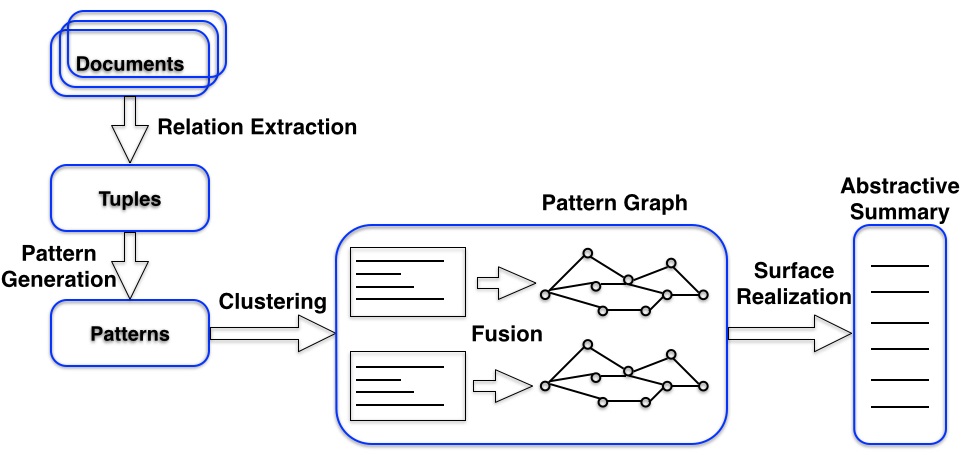
Figure 2: Document abstractive summarization framework flowchart.
This work was published in CIKM 2015.
Nugget 3. A new structured sparse learning model to integrate multi-dimensional features for risk prediction:
We propose a novel structured sparsity-inducing norms based feature learning model to integrate the multi-dimensional features for risk prediction. The new mixed norms are designed to learn the importance of different features from both local and global point of views. We successfully integrate multi-dimensional data to enhance classification results. The empirical results show that the proposed new method can effectively integrate different types of features, and consistently outperforms related methods using the concatenated feature vectors.
This work was published in KDD 2015.
Nugget 4. Discrete missing value imputation software:
In electronic medical records, there are many missing values. Especially many values are discrete, not continuous. Existing missing value imputation methods mainly focus on the continuous value prediction. In these cases, an additional step to process the continuous results with either heuristic threshold parameters or complicated mappings is necessary, while it is inefficient and may diverge from the optimal solution. To address this issue, we proposed a novel optimal discrete matrix completion model, which is able to learn the optimal thresholds automatically and also guarantees an exact low-rank structure of the target matrix. We derive stochastic gradient descent algorithm to optimize the new objective with proper strategies to speed up the optimization. In the experiments, it is proved that our method can predict discrete values with high accuracy, very close to or even better than these values obtained by carefully tuned thresholds. Meanwhile, our model is able to handle online data and easy to parallelize for big missing data imputation.
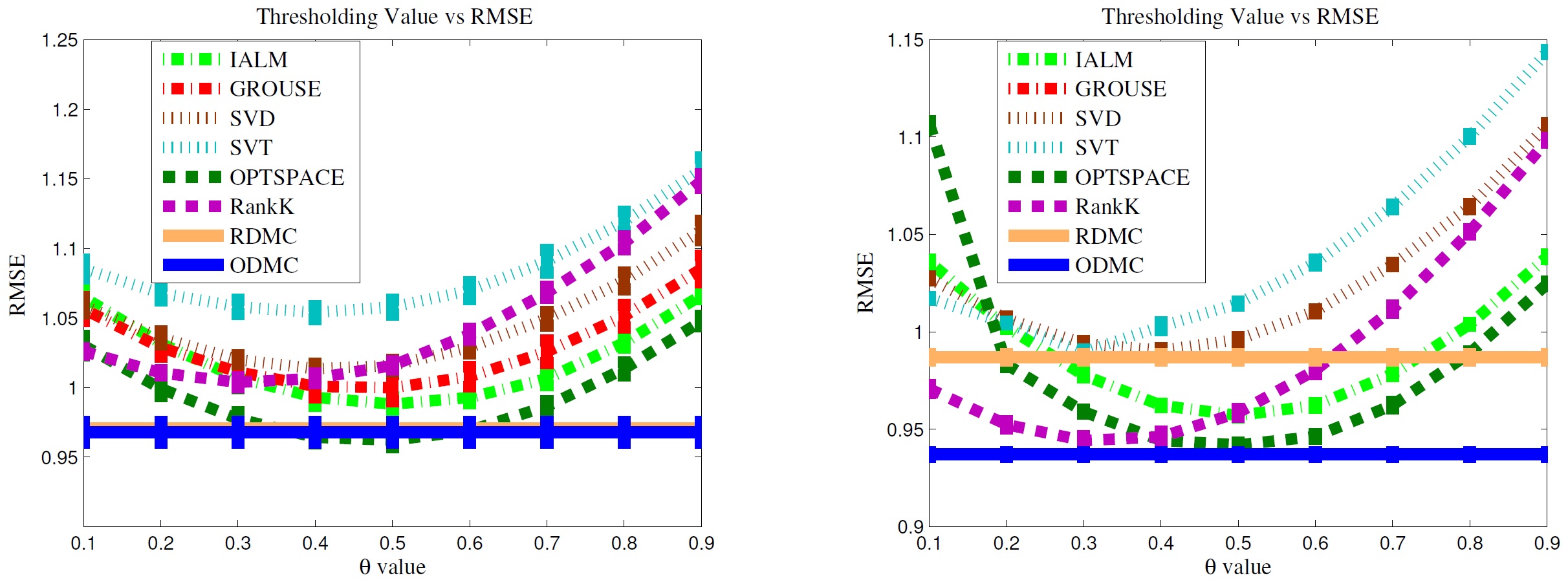
This work was published in AAAI 2016.
Some Related Publications:
Song Zhang, Jing Cao, Chul Ahn, Statistical Inference and Sample Size Calculation for Paired Binary Outcomes with Missing Data, 2016, Statistics in Medicine, in press.Y. Zhang, J. Cao, S. Zhang, A.J. Lee, G. Sun, C.N. Larsen, H. Zhao, Z. Gu, S. He, E.B. Klem, R.H. Scheuermann, Genetic changes found in a distinct clade of Enterovirus D68 associated with paralysis during the 2014 outbreak, Virus Evolution, 2016, 2(1): vew015.
Lei Du, Heng Huang, Jingwen Yan, Sungeun Kim, Shannon L. Risacher, Mark Inlow, Jason H. Moore, Andrew J. Saykin, Li Shen, ADNI. Structured Sparse Canonical Correlation Analysis for Brain Imaging Genetics: An Improved GraphNet Method. Bioinformatics, 32(10):1544-51.
De Wang, Feiping Nie, Heng Huang. Global Redundancy Minimization for Feature Ranking. IEEE Transactions on Knowledge and Data Engineering (TKDE), 27(10), pp. 2743-2755, 2015.
Peng Li, Heng Huang. Deep Learning Based Natural Language Processing System for Clinical Information Identification from Clinical Notes and Pathology Reports. The 10th International Workshop on Semantic Evaluation Joint with NAACL2016.
Zhouyuan Huo, Dinggang Shen, Heng Huang. New Multi-Task Learning Model to Predict Alzheimer's Disease Cognitive Assessment. 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2016), accepted to appear.
Xiaoqian Wang, Dinggang Shen, Heng Huang. Prediction of Memory Impairment with MRI Data: A Longitudinal Study of Alzheimer's Disease. 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2016), accepted to appear.
Xiaofeng Zhu, Heung-Il Suk, Heng Huang, Dinggang Shen. Structured Spare Low-Rank Regression Model for Brain-Wide and Genome-Wide Associations. 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2016), accepted to appear.
Xiaoqian Wang, Feiping Nie, Heng Huang. Structured Doubly Stochastic Matrix for Graph Based Clustering. 22nd ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD 2016), accepted to appear.
Zhouyuan Huo, Feiping Nie, Heng Huang. Robust and Effective Metric Learning Using Capped Trace Norm. 22nd ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD 2016), accepted to appear.
Wenhao Jiang, Hongchang Gao, Fu-Lai Korris Chung, Heng Huang. L2,1-Norm Stacked Robust Autoencoders for Domain Adaptation. Thirtieth AAAI Conference on Artificial Intelligence (AAAI 2016) , pp. 1723-1729.
Hua Wang, Cheng Deng, Hao Zhang, Xinbo Gao, Heng Huang. Learning Biological Relevance of Drosophila Embryos for Drosophila Gene Expression Pattern Annotations. Thirtieth AAAI Conference on Artificial Intelligence (AAAI 2016) , pp. 1324-1330.
Hongchang Gao, Feiping Nie, Heng Huang. Multi-View Subspace Clustering. International Conference on Computer Vision (ICCV 2015), pp. 4238-4246.
Hongchang Gao, Chengtao Cai, Jingwen Yan, Lin Yan, Joaquin Goni Cortes, Yang Wang, Feiping Nie, John West, Andrew Saykin, Li Shen, Heng Huang. Identifying Connectome Module Patterns via New Balanced Multi-Graph Normalized Cut. 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2015), pp. 169-176.
Yang Song, Weidong Cai, Heng Huang, Yun Zhou, Yue Wang, David Dagan Feng. Locality constrained Subcluster Representation Ensemble for Lung Image Classification. Medical Image Analysis (MIA), 22(1), pp. 102-113, 2015.
Hua Wang, Feiping Nie, Heng Huang. Large-Scale Cross-Language Web Page Classification via Dual Knowledge Transfer Using Fast Nonnegative Matrix Tri-Factorization. ACM Transactions on Knowledge Discovery from Data (TKDD), 10(1), pp. 1:1-1:29, 2015.
Yang Song, Weidong Cai, Heng Huang, Yun Zhou, David Dagan Feng, Yue Wang, Michael J. Fulham, Mei Chen. Large Margin Local Estimate with Applications to Medical Image Classification. IEEE Transactions on Medical Imaging (TMI), 34(6), pp. 1362-1377, 2015.
Hua Wang, Heng Huang, Chris Ding. Correlated Protein Function Prediction via Maximization of Data Knowledge Consistency. Journal of Computational Biology (JCB), 22(6), pp. 546-562, 2015.
Hongchang Gao, Lin Yan, Weidong Cai, Heng Huang. Anatomical Annotations for Drosophila Gene Expression Patterns via Multi-Dimensional Visual Descriptors Integration. 21th ACM SIGKDD Conference on Knowledge Discovery and Data Mining Conference (KDD 2015), pp. 339-348.
Xiaoqian Wang, Yun Liu, Feiping Nie, Heng Huang. Discriminative Unsupervised Dimensionality Reduction. Twenty-Fourth International Joint Conferences on Artificial Intelligence (IJCAI 2015), pp. 3925-3931.
Wenhao Jiang, Feiping Nie, Heng Huang. Robust Dictionary Learning with Capped L1 Norm. Twenty-Fourth International Joint Conferences on Artificial Intelligence (IJCAI 2015), pp. 3590-3596.
Yang Song, Weidong Cai, Qing Li, Dagan Feng, Heng Huang. Fusing Subcategory Probabilities for Texture Classification. Twenty-Eighth IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2015), pp. 4409-4417.
Shuai Zheng, Xiao Cai, Chris Ding, Feiping Nie, Heng Huang. A Closed Form Solution to Multiview Low-rank Regression. Twenty-Ninth AAAI Conference on Artificial Intelligence (AAAI 2015), pp. 1973-1979.
List of any software being distributed by project
Software 1: Structured sparse learning model to integrate multi-dimensional features for risk prediction:
Click to Download
Software 2: Discrete missing value imputation software:
Click to Download
Software 3: Robust metric learning using capped trace norm:
Click to Download
Software 4: Natural language processing based document abstractive summarization tool:
We are cleaning the code and will provide them soon.