This work introduces a novel and efficient algorithm for reconstructing
the 3D shapes of tumors from a set of 2D bioluminescence images which
are taken by the same camera but after continually rotating the animal
by a small angle. The method is efficient and robust enough to be
used for analyzing
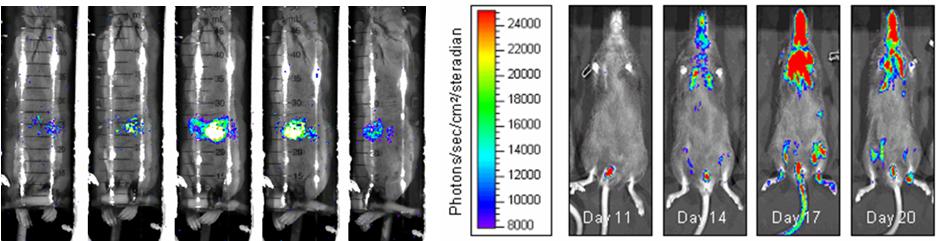
the repeated imaging of a same animal transplanted with gene marked
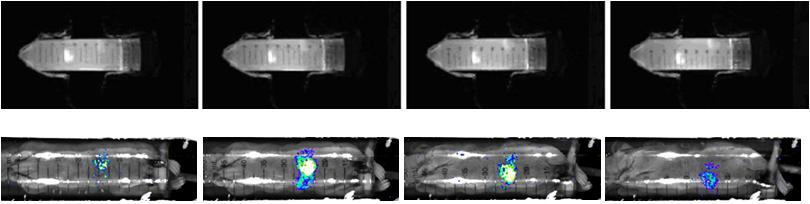
cells. There are several steps in our algorithm. First, the silhouettes
(or boundaries) of the animal and its interior hot spots (corresponding
to tumors) are segmented in the set of bioluminescence images. Second,
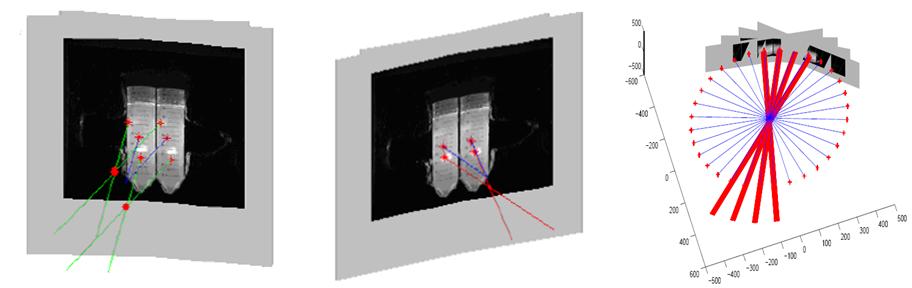
the images are registered according to the projection of the animal
rotating axis. Third, the images are mapped onto 3D projection planes
and from the viewpoint of each plane, the visual hulls of the animal
and its interior tumors are reconstructed. Then, the intersection
of visual hulls from all viewpoints approximates the shape of the
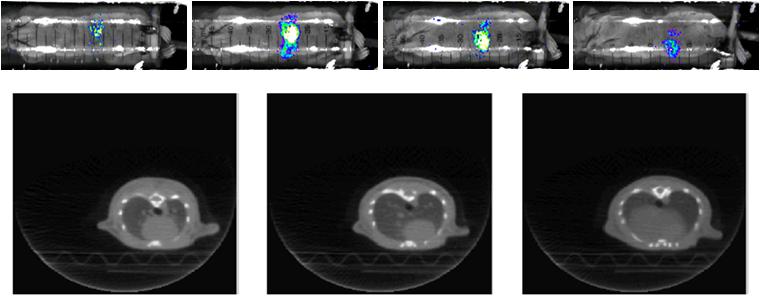
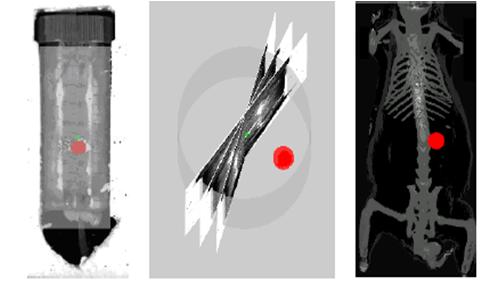
animal and its interior tumors. In order to visualize in 3D the structure
of the tumor, we also co-register the BLI-reconstructed crude structure
with detailed anatomical structure extracted from high-resolution
micro-CT on a single platform. The experimental results show promising
performance of our reconstruction and co-registration method.
.Papers
-
Junzhou Huang, Xiaolei Huang, Dimitris Metaxas, Debarata Banerjee, ”3D Tumor Shape Reconstruction from 2D Bioluminescence Images and Registration with CT Images”, 1st Workshop on Microscopic Image Analysis with Applications in Biology, MIAAB’06, 2006. (Oral)
-
Junzhou Huang, Xiaolei Huang, Dimitris Metaxas, Debarata Banerjee, ”3D Tumor Shape Reconstruction from 2D Bioluminescence Images” , IEEE Int’l Symposium on Biomedical Imaging: From Nano to Macro, ISBI’06, pp. 606-609, 2006. (Oral)
Bioluminense Images
 |
|
Samples
 |
|
Tumor Localization, Segmentation, Reconstruction
 |
|
Registration bewtween Bioluminense Images and CT images
 |
|
Final Visualization Results
 |
|