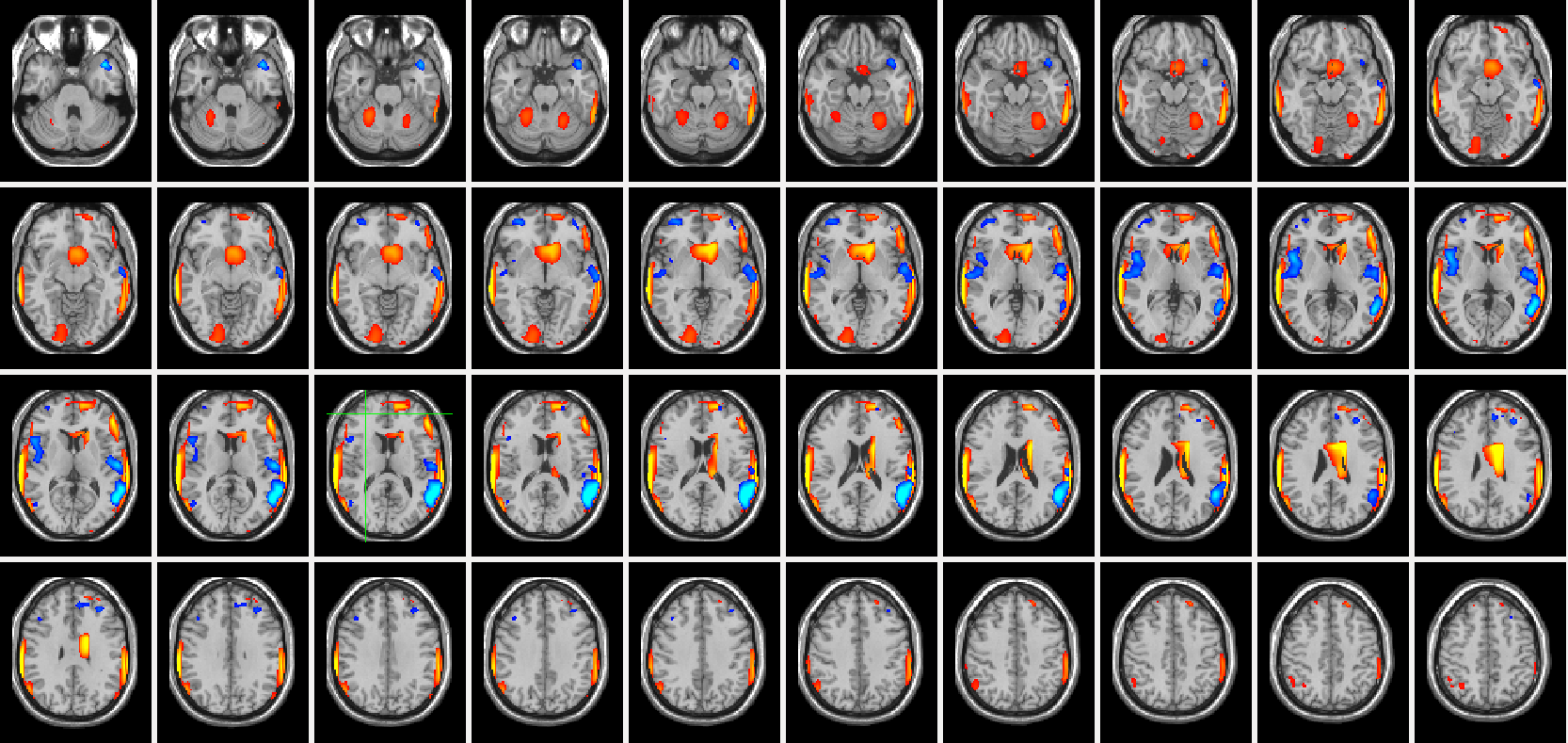
Statistical analysis of longitudinal or cross sectional brain imaging data to identify effects of neurodegenerative diseases is a fundamental task in various studies in neuroscience. However, when there are systematic variations in the images due to parameter changes such as changes in the scanner protocol, hardware changes, or when combining data from multi-site studies, the statistical analysis becomes problematic. Motivated by this scenario, the goal of this paper is to develop a unified statistical solution to the problem of systematic variations in statistical image analysis. Based in part on recent literature in harmonic analysis on diffusion maps, we propose an algorithm which compares operators that are resilient to the systematic variations. These operators are derived from the empirical measurements of the image data and provide an efficient surrogate to capturing the actual changes across images. We also establish a connection between our method to the design of wavelets in non-Euclidean space. To evaluate the proposed ideas, we present various experimental results on detecting changes in simulations as well as show how the method offers improved statistical power in the analysis of real longitudinal PIB-PET imaging data acquired from participants at risk for Alzheimer’s disease (AD).
NIH R01AG040396, NIH R01AG021155, NIH AG037639, and NSF CAREER Award RI1252725. Partial support was provided by UW ADRC (AG033514), UW ICTR (1UL1RR025011), UW CPCP (AI117924) and NIH grants AG010129 and AG027161.